A new method proposed by University of Guelph researchers that uses artificial intelligence to analyze ultrasound images could one day deliver big results for little ones.
The AI method they propose could lead to faster and more accurate diagnoses of serious brain problems in newborn and premature infants.
About 20-30 per cent of low-weight babies can develop hemorrhaging in a part of the brain called the ventricles, which are fluid-filled cavities that keep the brain buoyant and cushioned. Left undiagnosed, hemorrhaging can lead to lifelong brain damage.
One tool for diagnosing the problem, 2D ultrasound, creates images that are often blurry and grainy. 3D ultrasound is better, but the images are difficult and time-consuming to analyze manually.
Researchers from the College of Engineering and Physical Sciences have found that employing an AI method called semi-supervised learning (SSL) can segment and analyze these grainy ultrasound images to clarify the shape and size of the brain’s ventricles, helping doctors determine the severity of hemorrhaging.
The researchers, led by Dr. Eranga Ukwatta and PhD candidate Zachary Szentimrey, say this method could help doctors make accurate diagnoses to begin interventions faster.
“This research has the potential to significantly improve the way we diagnose conditions in newborns,” says Ukwatta. “By using AI to analyze ultrasound images, we can get faster and more accurate results without subjecting the baby to uncomfortable or potentially risky procedures. This can lead to earlier intervention and better outcomes for these fragile patients.”
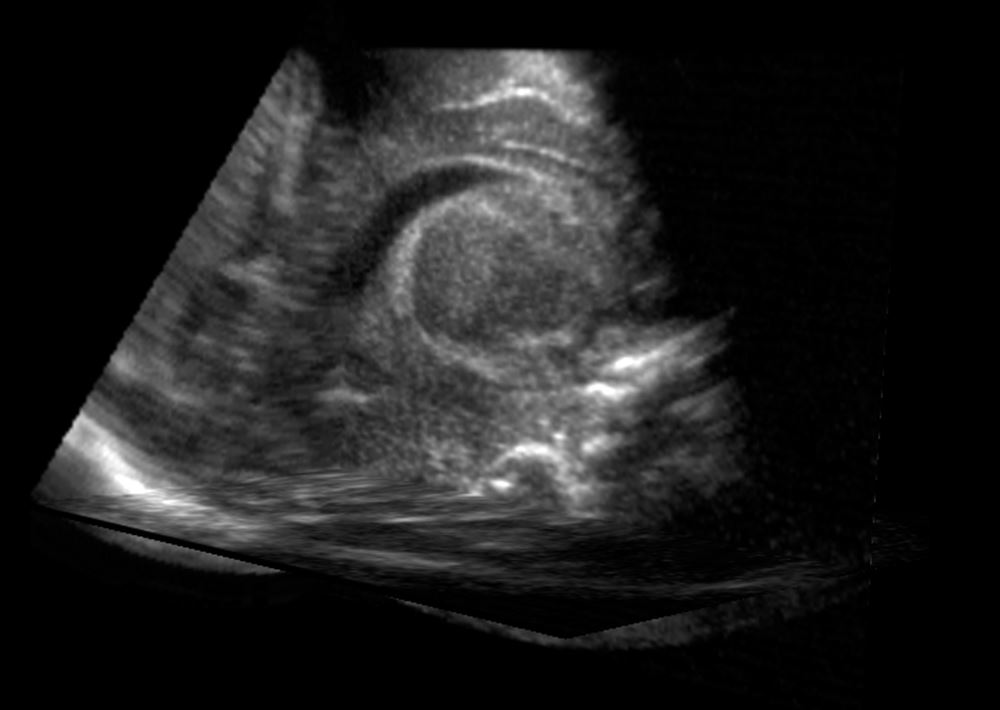
Ultrasound image
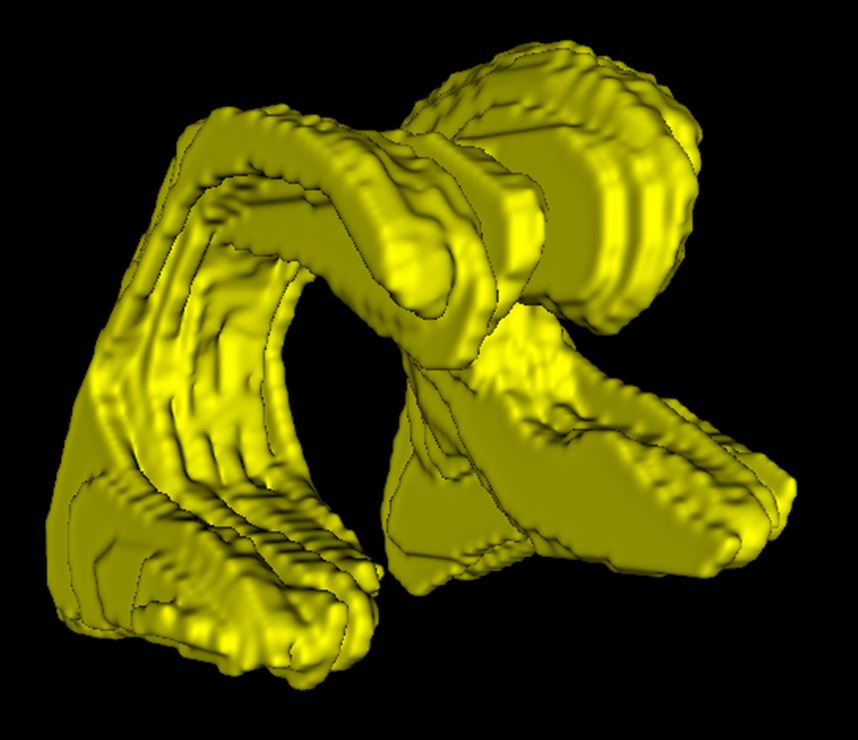
Manual analysis of ultrasound
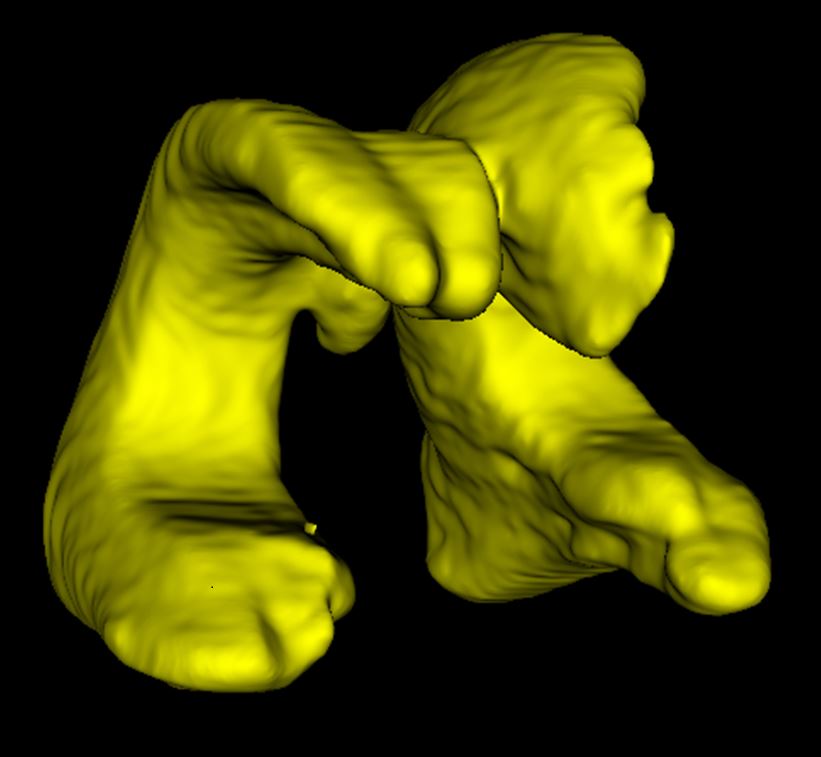
AI analysis of ultrasound
When analyzing the brain’s ventricles, manual and AI analyses of the ultrasounds were comparable with one another. AI may be a more practical option for clinics.
New AI method, ‘semi-supervised learning,’ helps avoid risky MRI and CT scans
The use of SSL for analyzing 3D ultrasounds was detailed in a study recently published in Medical Physics. It is one of the first studies to apply this special learning method to organ structure and 3D ultrasounds.

Here’s how it works:
First, the AI is trained on a small number of ultrasounds that have already been analyzed by doctors, to teach the AI the basic shapes and sizes of healthy and non-healthy ventricles.
Then, the AI is shown unanalyzed ultrasounds. By comparing blurry images to what it learned earlier, the AI can make an accurate guess about the shape and size of the ventricles in new images.
This AI method was found to outperform traditional analysis methods in both accuracy and speed. In the study, it also achieved higher accuracy in identifying and segmenting the brain’s ventricles with fewer computational resources.
Using this method would allow doctors to avoid other diagnostic tools such as magnetic resonance imaging (MRI) and computed tomography (CT) scans, which can be uncomfortable for babies and, in the case of CT scans, can carry health risks due to radiation exposure.
The new AI method could be a more practical option for clinical settings. It would provide a non-invasive, efficient and accurate way to monitor and diagnose potential brain issues in newborns, potentially leading to quicker and more effective medical interventions.
“This is just the beginning,” Szentimrey says. “We’re also exploring ways to improve the AI so it can analyze images taken by less experienced medical professionals or even technicians in remote locations. This could make this technology even more accessible and beneficial for neonatal intensive care units around the world.”
U of G’s expanding AI innovations
This project builds on U of G’s growing innovations in AI research and technologies. This includes the Centre for Advancing Responsible and Ethical Artificial Intelligence (CARE-AI), which focuses on applying machine learning and AI to U of G’s research strengths, including human and animal health, environmental sciences, agriculture, business and the bio-economy.
Additionally, computer science professor Dr. Rozita Dara recently launched Artificial Intelligence for Food (AI4Food), a collaborative research initiative from CEPS to improve resilience, safety, production, processing and sustainability in national and international agri-food systems.
Contact:
Dr. Eranga Ukwatta
eukwatta@uoguelph.ca
This study was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (EU), AMSOSO and the Canadian Institutes of Health Research (CIHR). Computations were performed on the Niagara supercomputer at the SciNet HPC Consortium. SciNet is funded by the Canada Foundation for Innovation; the Government of Ontario; Ontario Research Fund—Research Excellence and the University of Toronto.
This story was written by Mojtaba Safdari as part of the Science Communicators: Research @ CEPS initiative. Mojtaba is a PhD candidate in the School of Engineering under Dr. Amir A. Aliabadi.
This study was done in collaboration with Dr. Aaron Fenster at the Western University, where the data was accessible to U of G researchers through a data sharing agreement.
